Scientific Workflow Management
This page is work in progress.
This module provides an overview on how a CHESS can use notion of workflows to automate their data processing tasks and in the process shortening the turn around time for the data processing that needs to be done on data collected at a beamline.
The content in this module is gathered from author’s various presentations over the years and their group’s work on workflows.
What are Scientific Workflows
Scientific workflows allow users to easily express multi-step computational tasks, for example retrieve data from an instrument or a database, reformat the data, and run an analysis. A scientific workflow describes the dependencies between the tasks and in most cases the workflow is described as a directed acyclic graph (DAG), where the nodes are tasks and the edges denote the task dependencies.
A defining property for a scientific workflow is that it manages data flow. The tasks in a scientific workflow can be everything from short serial tasks to very large parallel tasks (MPI for example) surrounded by a large number of small, serial tasks used for pre- and post-processing.
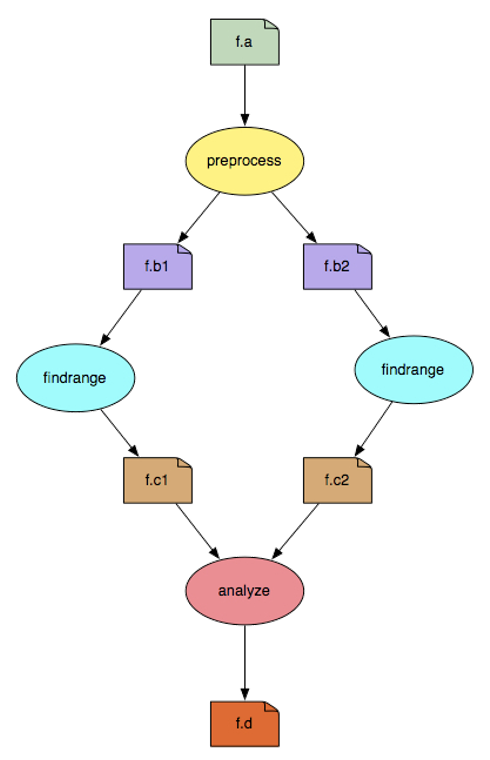
In the above example figure, the nodes/vertices in indicate the job that needs to be run, while the edges indicate the dependencies between the jobs.
The dependencies can be both control flow or data driven i.e. - a job can be run only after the parent has finished successfully OR - a job requires data sets to run that are generated as outputs by a parent node/job. - the rectangles in the figure indicate datasets that a node/job require and generate.
Using scientific workflows for your data processing in general provides you the following benefits
Reproducibility - Scientific workflows allow you to document and reproduce your analyses, ensuring their validity.
Automation - Workflows automate repetitive and time-consuming tasks, thereby reducing the workload of researchers.
Scalability - Workflows allow you to scale up your data processing to handle large data sets and complex analyses, enabling you to solve large research problems in your field. It helps you run your analysis in parallel over distributed resources.
Reusability - Once your data processing pipeline is modelled as a workflow, typically it is possible to reuse the workflow in different ways. For example, you could use the workflow as part of a bigger science analysis (that maybe faclity wide), or even share the workflow with another CHESS researcher.
High Level Steps on Identifying a Workflow
Here are some tips on how you can identify a workflow for your data processing
- Start on a whiteboard and think about the various steps that your data processing entails. Especially when you already have existing monolithic code for data processing, it is important to think about your processing at a high level in a logical manner.
- Think about the various steps that make sense logically, and would also make sense another CHESS researcher when discussing the problem.
- Think about the granularity in context of input data. If a particular step in your identified workflow, can benefit on working on smaller chunks of data; then consider breaking down that step in smaller parallel sub steps. For example if you are working on time series dataset, and you have a dataset collected for a month, you can break down processing in chunks of
- per day
- per hour
- per minute
- and so forth.
- Often, each step (type of step) in your workflow would map to a different executable.
Workflow Constructs
There are some core different types of workflow patterns that underpin most of the complex workflows. The jobs in the examples, below map to simple commonly available linux executables and is for illustration purposes only. For your actual workflows, you use your actual scientific code.
Process

In the above example, workflow consists of a single job that runs the ls command and generates a listing of the files in the / directory.
Pipeline
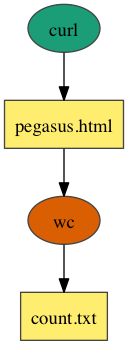
In the above example, workflow consists of two jobs linked together in a pipeline. The first job runs the curl command to fetch the Pegasus home page and store it as an HTML file. The result is passed to the wc command, which counts the number of lines in the HTML file.
Split
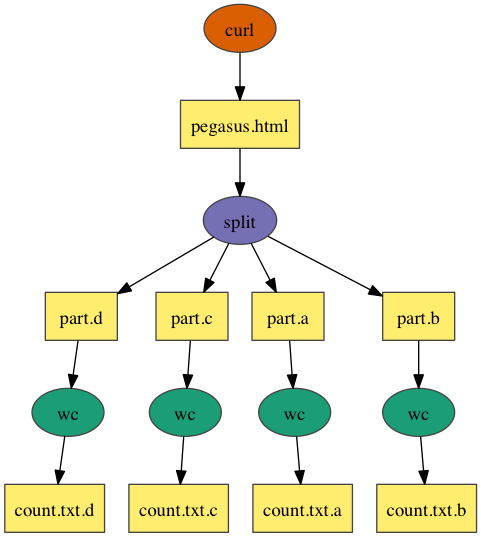
In the above example, workflow consists of a job that downloads the Pegasus home page using the curl command, then uses the split command to divide it into 4 pieces. The result is passed to the wc command to count the number of lines in each piece.
Merge
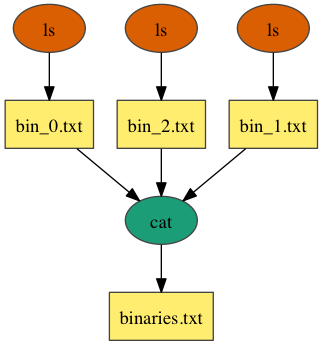
In the above example, workflow consists of 3 jobns that execute the ls command on several */bin directories and passes the results to the cat command, which merges the files into a single listing.
Workflow Challenges
When running workflows independent of what workflow system you are using, there are some common challenges that you may encounter.
- How to describe your complex workflows in a simple way? This is important in relation to sharing your workflows with your colleagues. Can the workflow that you just ran locally on your resource, can be run by another researcher on the
- same resource
- different resource / cluster
- what changes if any are required?
- How do you get your workflows to access distributed, heterogeneous data and resources (heterogeneous interfaces). For example, you may want to distribute your analysis across different clusters.
- How to deal with resources/software that change over time? The resource on which you are currently running may go away (no longer operational), software dependencies on which your code requires change.
- How to have ease of use? Ability to debug and monitor large workflows.
Workflow Creation
There are a variety of workflow systems available that you can use in general. Some of them allow you to create workflows using
- A graphical interface
- Programmatically using an API in a main stream programming language such as Python
CHESS researchers have access to the following systems for running workflows
- CHESS Workflow Runner
- Galaxy (to be covered in DC102)
- Pegasus Workflows
Pegasus Workflows
Pegasus WMS allows users to model their computational pipelines as workflows, that can execute in a number of different environments including
- desktops,
- campus clusters,
- distributed computing environments such as PATh and OSG OSPool,
- supercomputing CI such as ACCESS and clouds.
Pegasus bridges the scientific domain and the execution environment by automatically mapping high-level workflow descriptions onto distributed resources. It automatically locates the necessary input data and computational resources necessary for workflow execution. Pegasus enables scientists to construct workflows in abstract terms without worrying about the details of the underlying execution environment or the particulars of the low-level specifications required by the middleware (HTCondor, SLURM, or Amazon EC2).
The clean separation of how the user defines a Pegasus workflow (in a resource agnostic way) called the Abstract workflow and the Executable workflow that actually runs on your target resource is illustrated below.
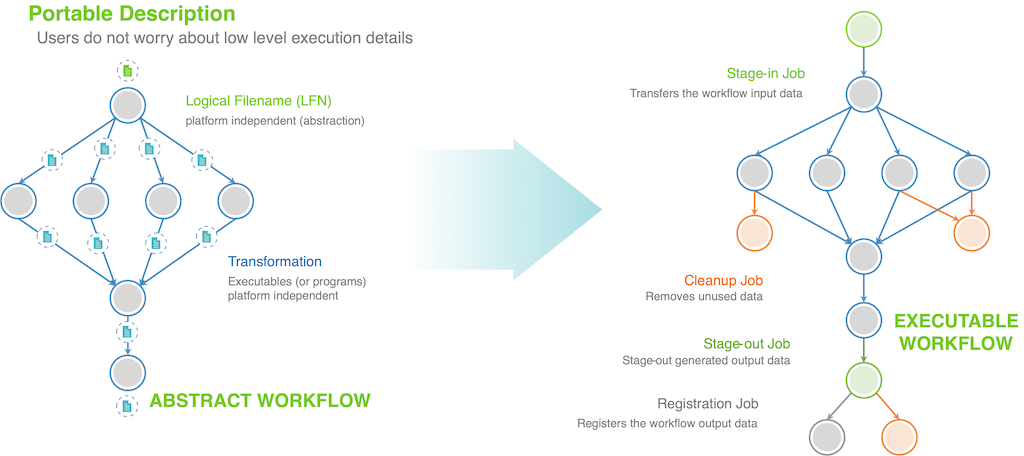
As a user when you define a Pegasus workflow, you
- identify both input and data sets that a job requires by their logical identifiers called LFN.
- identify the executable that needs to be invoked for a job by it’s logical identifier called transformation.
The Abstract workflow when given to Pegasus gets transformed to an Executable workflow that can execute on your target resource. As part of this transformation of the workflow Pegasus will figure out
- how to get/stage the input data required for your workflow. add data stage-in jobs
- maps your jobs to a compute resource
- how to stage-out data that your workflow generates
Additionally, as part of this transformation Pegasus can do a lot of optimizations on your workflow such as
- add data cleanup nodes, that clean up data from cluster when it is not required
- cluster short running jobs together
- data reuse (delete jobs whose datasets already exist)
Getting Started with Pegasus @ CHESS
Pegasus is already installed and configured to run on the CHESS cluster. In order to use Pegasus, you need to login to the node lnx201.classe.cornell.edu .
X-CITE vahi$ ssh lnx201.classe.cornell.edu
(user@lnx201.classe.cornell.edu) Password: Source Pegasus binaries in your PATH
In order to run Pegasus you need to make sure that Pegasus executables are in your PATH. You can do this by sourcing the following setup file.
[kvahi@lnx201 ~]$ source /nfs/chess/sw/pegasus/setup.sh
(pegasus-env) [kvahi@lnx201 ~]$ pegasus-version
5.1.0devSetup a test diamond workflow and run it
In this example, we will try and run a test diamond workflow through Pegasus.
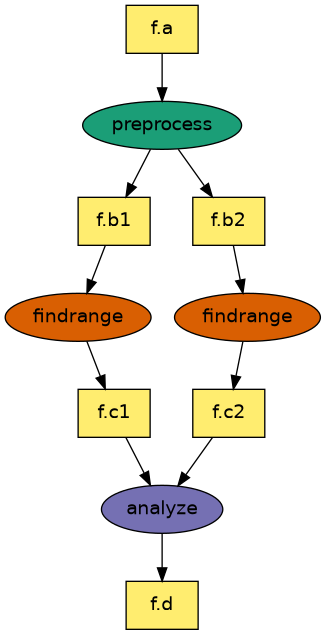
To setup a test workflow that runs on the CHESS SGE cluster, you can use the pegasus-init executable and answer the questions asked.
You should do the following selections when prompted
- Select an execution environment [1]: 5
- What’s the execution environment’s queue: chess.q
- What’s your project’s name []:
- Select an example workflow [1]: 1
[kvahi@lnx201 pegasus]$ pegasus-init diamond
###########################################################
########### Available Execution Environments ##########
###########################################################
1) Local Machine Condor Pool
2) Local SLURM Cluster
3) Remote SLURM Cluster
4) Local LSF Cluster
5) Local SGE Cluster
6) OLCF Summit from OLCF Headnode
7) OLCF Summit from OLCF Hosted Kubernetes Pod
Select an execution environment [1]: 5
What's the execution environment's queue: chess.q
What's your project's name []:
What's the location of the PEGASUS_HOME dir on the compute nodes [/nfs/chess/user/kvahi/software/pegasus/default]:
###########################################################
########### Available Workflow Examples ##########
###########################################################
1) pegasus-isi/diamond-workflow
2) pegasus-isi/hierarichal-sample-wf
3) pegasus-isi/merge-workflow
4) pegasus-isi/pipeline-workflow
5) pegasus-isi/split-workflow
Select an example workflow [1]: 1
Fetching workflow from https://github.com/pegasus-isi/diamond-workflow.git
Generating workflow based on pegasus-isi/diamond-workflow
This workflow will target queue "chess.q"
The PEGASUS_HOME location is "/nfs/chess/user/kvahi/software/pegasus/default"
Creating workflow properties...
Creating transformation catalog...
Creating replica catalog...
Creating diamond workflow dag...
INFO:Pegasus.api.workflow:diamond added Job(_id=ID0000001, transformation=preprocess)
INFO:Pegasus.api.workflow:diamond added Job(_id=ID0000002, transformation=findrange)
INFO:Pegasus.api.workflow:diamond added Job(_id=ID0000003, transformation=findrange)
INFO:Pegasus.api.workflow:diamond added Job(_id=ID0000004, transformation=analyze)
INFO:Pegasus.api.workflow:inferring diamond dependencies
INFO:Pegasus.api.workflow:workflow diamond with 4 jobs generated and written to workflow.yml
Creating properties file...
Creating site catalog for SitesAvailable.SGE...The example workflow gets generated in the diamond directory.
[kvahi@lnx201 pegasus]$ cd diamond/
(pegasus-env) [kvahi@lnx201 diamond]$ ls
diamond-workflow generate.sh pegasus.properties plan.sh replicas.yml sites.yml transformations.yml workflow.ymlIn this directory, you will see the following yaml files
- workflow.yml - the abstract workflow generated for the diamond workflow that we will run.
- replicas.yml - a yaml formatted file that lists the locations of the inputs for the workflow.
- transformations.yml - a yaml formatted file that lists the locations of executables used by the workflows.
- sites.yml - a yaml formated file that describe the CHESS SGE cluster to Pegasus.
The above catalog files are described in the Pegasus documentation.
To plan the workflow, run the ./plan.sh script.
(pegasus-env) [kvahi@lnx201 diamond]$ ./plan.sh
./plan.sh
2025.05.13 14:56:10.755 EDT:
2025.05.13 14:56:10.761 EDT: -----------------------------------------------------------------------
2025.05.13 14:56:10.766 EDT: File for submitting this DAG to HTCondor : diamond-0.dag.condor.sub
2025.05.13 14:56:10.771 EDT: Log of DAGMan debugging messages : diamond-0.dag.dagman.out
2025.05.13 14:56:10.777 EDT: Log of HTCondor library output : diamond-0.dag.lib.out
2025.05.13 14:56:10.782 EDT: Log of HTCondor library error messages : diamond-0.dag.lib.err
2025.05.13 14:56:10.787 EDT: Log of the life of condor_dagman itself : diamond-0.dag.dagman.log
2025.05.13 14:56:10.792 EDT:
2025.05.13 14:56:10.798 EDT: -no_submit given, not submitting DAG to HTCondor. You can do this with:
2025.05.13 14:56:10.808 EDT: -----------------------------------------------------------------------
2025.05.13 14:56:23.174 EDT: Database version: '5.1.0dev' (sqlite:////home/kvahi/.pegasus/workflow.db)
2025.05.13 14:56:28.480 EDT: Pegasus database was successfully created.
2025.05.13 14:56:28.485 EDT: Database version: '5.1.0dev' (sqlite:////nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001/diamond-0.replicas.db)
2025.05.13 14:56:28.693 EDT: Output replica catalog set to jdbc:sqlite:/nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001/diamond-0.replicas.db
2025.05.13 14:56:28.693 EDT:
I have concretized your abstract workflow. The workflow has been entered
into the workflow database with a state of "planned". The next step is
to start or execute your workflow. The invocation required is
pegasus-run /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
2025.05.13 14:56:29.864 EDT: Time taken to execute is 28.297 seconds The above command generated the executable workflow that you can run.
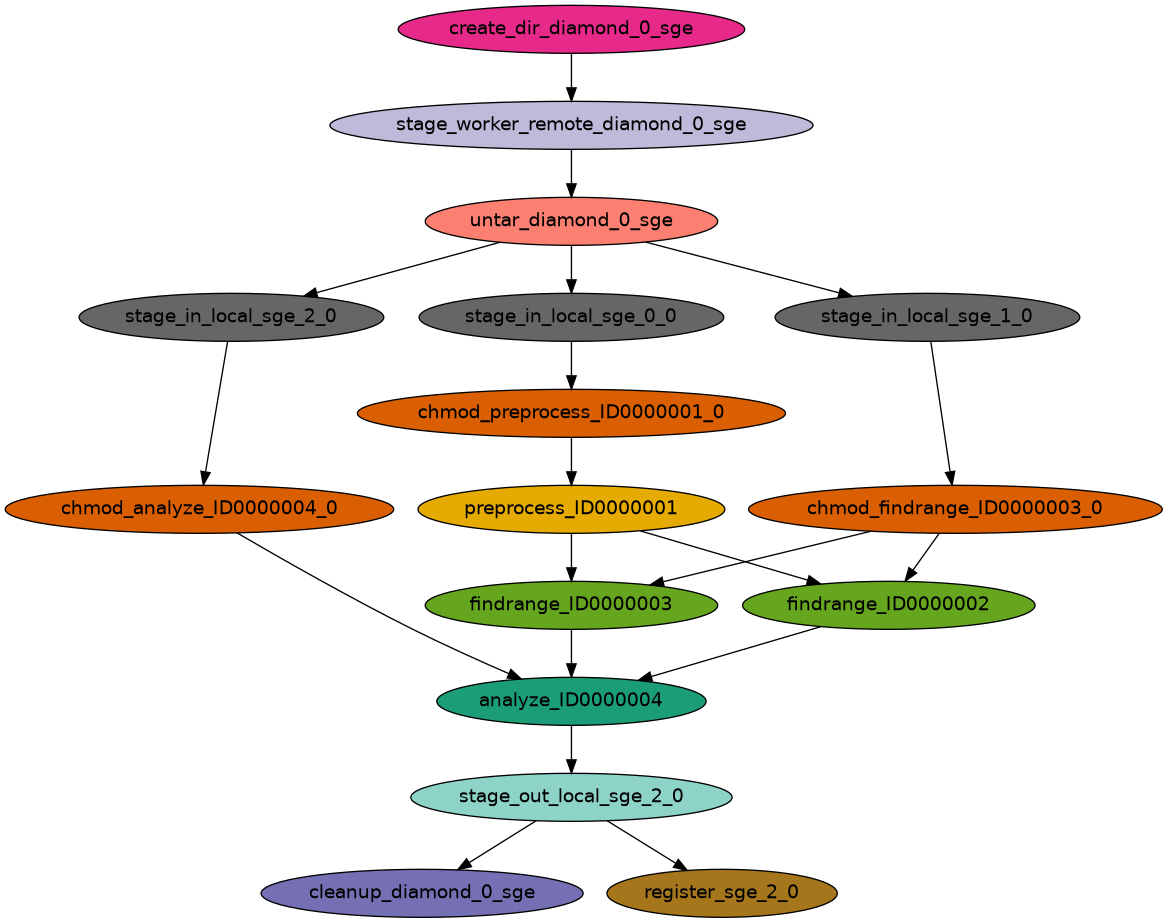
To do this copy the pegasus-run command invocation that you see in your terminal.
(pegasus-env) [kvahi@lnx201 diamond]$ pegasus-run /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
Submitting to condor diamond-0.dag.condor.sub
Submitting job(s).
1 job(s) submitted to cluster 693.
Your workflow has been started and is running in the base directory:
/nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
*** To monitor the workflow you can run ***
pegasus-status -l /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
*** To remove your workflow run ***
pegasus-remove /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001To check the status of the worklfow we can run the command pegasus-status with a -w 30 option to watch it every 30 seconds.
Again make sure you copy the pegasus-status invocation that you see in your terminal and add the -w 30 option after -l option.
(pegasus-env) [kvahi@lnx201 diamond]$ pegasus-status -l -w 30 /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
ID SITE STAT IN_STATE JOB
693 local Run 03:06 diamond-0 (/nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001)
696 sge Done 00:04 ┗━untar_diamond_0_sge
Summary: 2 Condor jobs total (R:1)
UNREADY READY PRE IN_Q POST DONE FAIL %DONE STATE DAGNAME
13 0 0 0 1 2 0 12.5 Running diamond-0.dag Once the workflow completes you will see somthing similar to the following
pegasus-env) [kvahi@lnx201 diamond]$ pegasus-status -l -w 30 /nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001
Press Ctrl+C to exit (pid=2380412) Tue May-13-2025 15:07:51
ID SITE STAT IN_STATE JOB
693 local Run 08:58 diamond-0 (/nfs/chess/user/kvahi/diamond/submit/kvahi/pegasus/diamond/run0001)
Summary: 1 Condor job total (R:1)
UNREADY READY PRE IN_Q POST DONE FAIL %DONE STATE DAGNAME
0 0 0 0 0 16 0 100.0 Success diamond-0.dag
Summary: 1 DAG total (Success:1)You can also run additional pegasus commands after your worklfow finishes. The
- pegasus-analyser
- This allows you to debug your workflow if it fails. Will tell you what jobs failed and why. - pegasus-statistics
- This allows you to generate runtime statistics about your workflow run such how long it run, how much resources were used etc.
Support
If you would like to use Pegasus to run your pipeline you can contact via
- Email - Support requests and bug reports can be emailed to pegasus-support@isi.edu.
- Slack - We encourage you to join the Slack Workspace as it is an
on-going, open forum for all Pegasus users to share ideas, experiences, and talk out issues with the Pegasus Development team. Please ask for an invite by trying to join pegasus-users.slack.com in the Slack app, or email pegasus-support@isi.edu and request an invite.
CHESS Workflow Runner
To be added. Coming soon in FALL 2025.